A mathematical model could help understand the mechanical force behind protein synthesis.
Researchers find features that shape mechanical force during protein synthesis
Posted on March 4, 2019UNIVERSITY PARK, Pa. — Like any assembly line, the body’s protein-building process generates a mechanical force as it produces these important cellular building blocks. Now, a team of researchers suggest they are one step closer to understanding that force. They also built a mathematical model to help guide scientists with future investigations into how the body creates proteins.
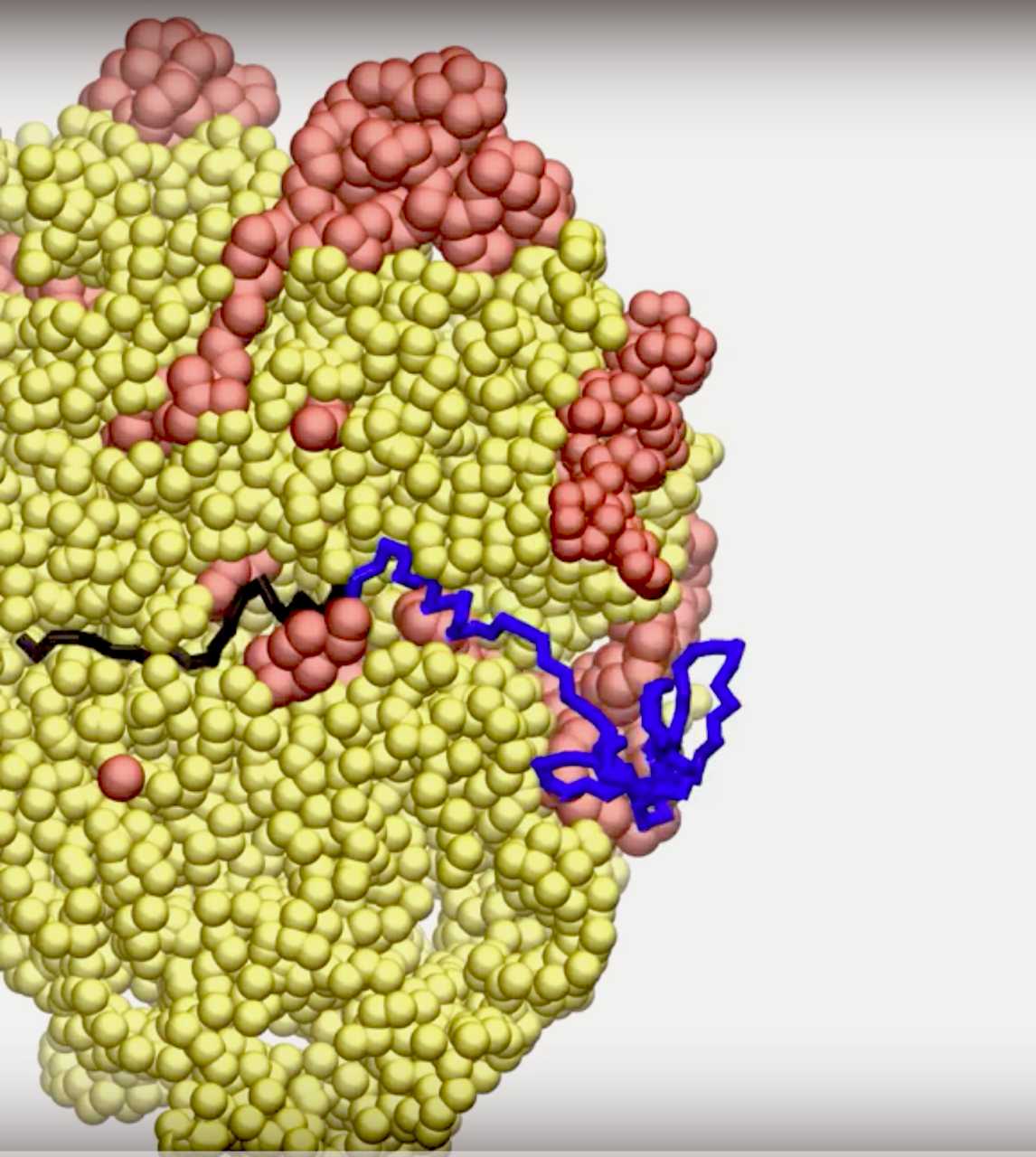
“In the past five years, it has been found experimentally that when a protein self-assembles and folds into its three-dimensional structure during its synthesis, a mechanical force can be generated that changes the speed of protein synthesis,” said Edward O’Brien, assistant professor of chemistry at Penn State and an Institute for Computational and Data Sciences co-hire. “What was unknown until this study is what features of protein folding determine the strength of that mechanical force. It’s important to understand such factors that influence the speed at which this machine works because the speed has been shown to determine protein function, structure and behavior in cells.”In this image, a protein (blue and black) is beginning to make its long (molecularly speaking) journey from the ribosome (red and yellow) through the tube and toward its eventual folding. IMAGE: Penn State
Proteins, which are used to make tissues, bones and muscles, are synthesized by the ribosome, a molecular machine whose purpose is to convert the genetic information contained in messenger RNA, or mRNA, into a protein. The ribosome moves along a mRNA, reading the code it contains and adding links to the elongating protein chain, which exits the ribosome through a narrow tube. Eventually, this process stitches together an unfolded protein and, once it exits the tube, it begins the complex folding process that leads to its three-dimensional shape.
According to O’Brien, the shape — or topology — of the protein, and its stability, as well as the speed of translation, determine the mechanical force that the folding process generates. He added that the distance traveled by the nascent protein during the process is actually quite large, at least compared to the dimensions of the minute world of chemical reactions.
“As the protein is being synthesized, it emerges through a narrow channel, or tube, which is about 10 nanometers in length and 1.5 nanometers in diameter,” said O’Brien. “The protein doesn’t fold within that tube, but it begins to fold as the nascent chain emerges out of that tube. What’s interesting to us, and to the community, is that this force is transmitted over 10 nanometers, which is long compared to molecular length scales. The length of a chemical bond, in comparison, is about .1 nanometers.”
This mechanical force can then provide feedback and change the speed at which synthesis happens, said O’Brien.
Sarah Leininger, doctoral candidate in chemistry at Penn State and first author of the paper, said the team used considerable computer resources for the study, which was published online today (March 1) in the Proceedings of the National Academy of Sciences. She added that the team used about 5 million computer hours to get the results for the study and create simulations, which resemble movies, of the synthesis process.
O’Brien said that because of the amount of computational power needed to simulate the complexity of protein synthesis, the team used a simulation model that helped them make accurate approximations of the process.
“These simulations were primarily molecular dynamic simulations that allow us to model the evolution of the system over time,” said O’Brien. “The key aspect of our model is we use coarse-grained simulations, where we don’t represent every single atom, but we approximate and describe groups of atoms, grouping them into one interaction site in our simulations. That lets us simulate longer time scales and be able to model this process that we wouldn’t be able to model otherwise.”
The simulations — and the equation devised by the researchers that describes the simulations — could help save time and money for scientists studying protein synthesis. Rather than conducting expensive experiments, or months-long simulations on a supercomputer, for example, scientists could perform calculations on a laptop computer in seconds.
“What this allows us to do — and other scientists to do — is without doing expensive simulations, without even doing experiments, they can reasonably estimate what will happen,” said O’Brien.
Fabio Trovato and Daniel Nissley, both postdoctoral scholars in chemistry, worked with O’Brien and Leininger.
The research was performed, in part, using resources and the computing assistance of Penn State’s Institute for CyberScience Advanced CyberInfrastructure (ICDS-ACI).
